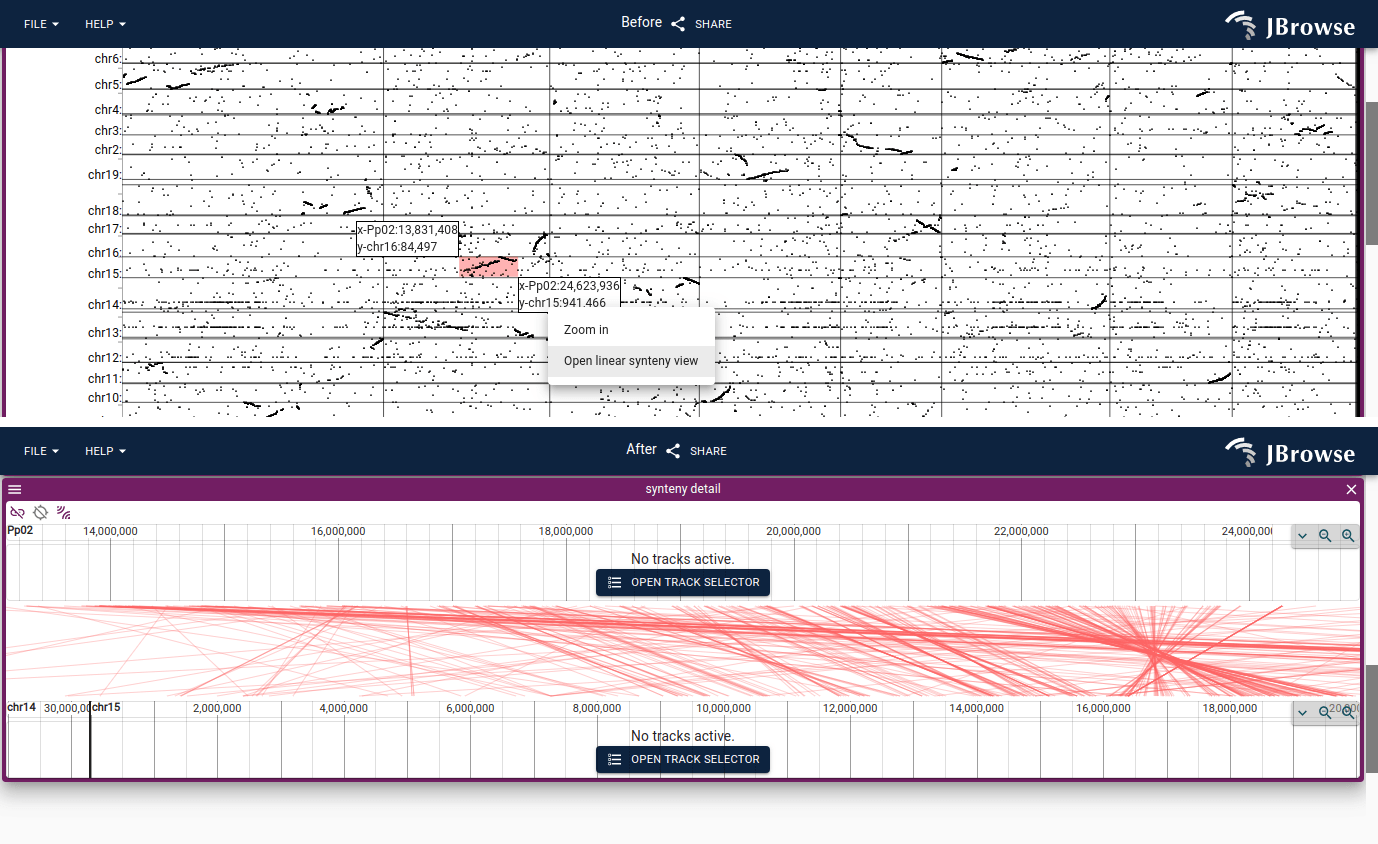
Dotplot view
The dotplot view is a 2D comparative view that can display alignments between different genome assemblies, or even compare a long-read or NGS short-read against the genome.
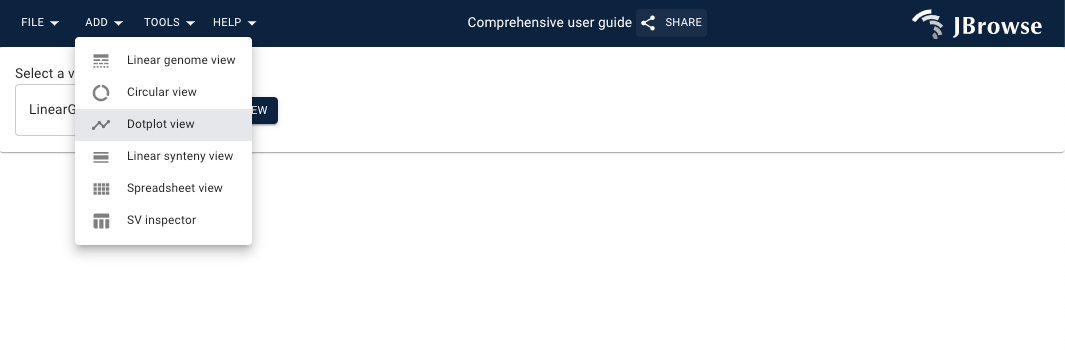
Opening a dotplot view
- Navigate on the header bar
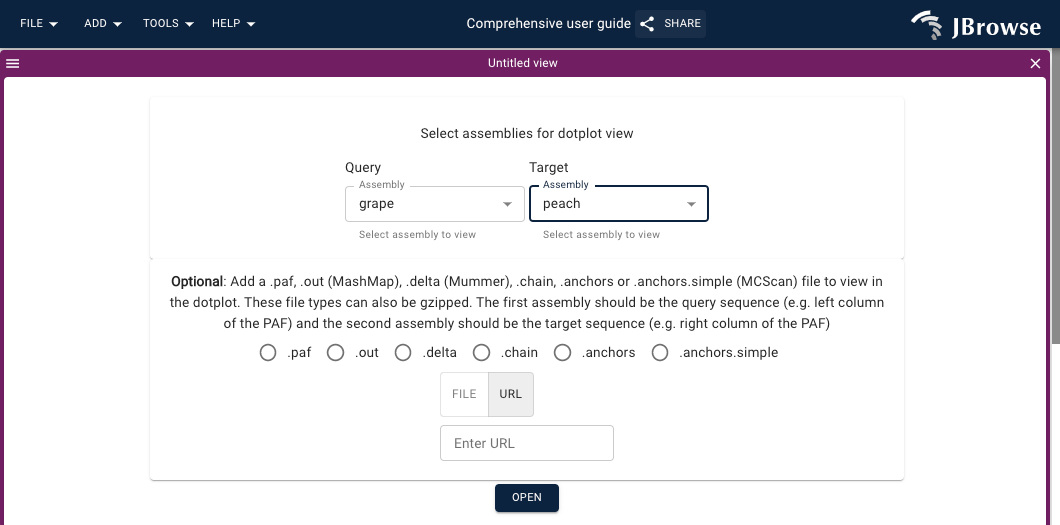
Add->Dotplot view - Select the genome assemblies of interest
- Optionally, add a .paf, .out (MashMap), .delta (Mummer), .chain, .anchors or .anchors.simple (MCScan) file
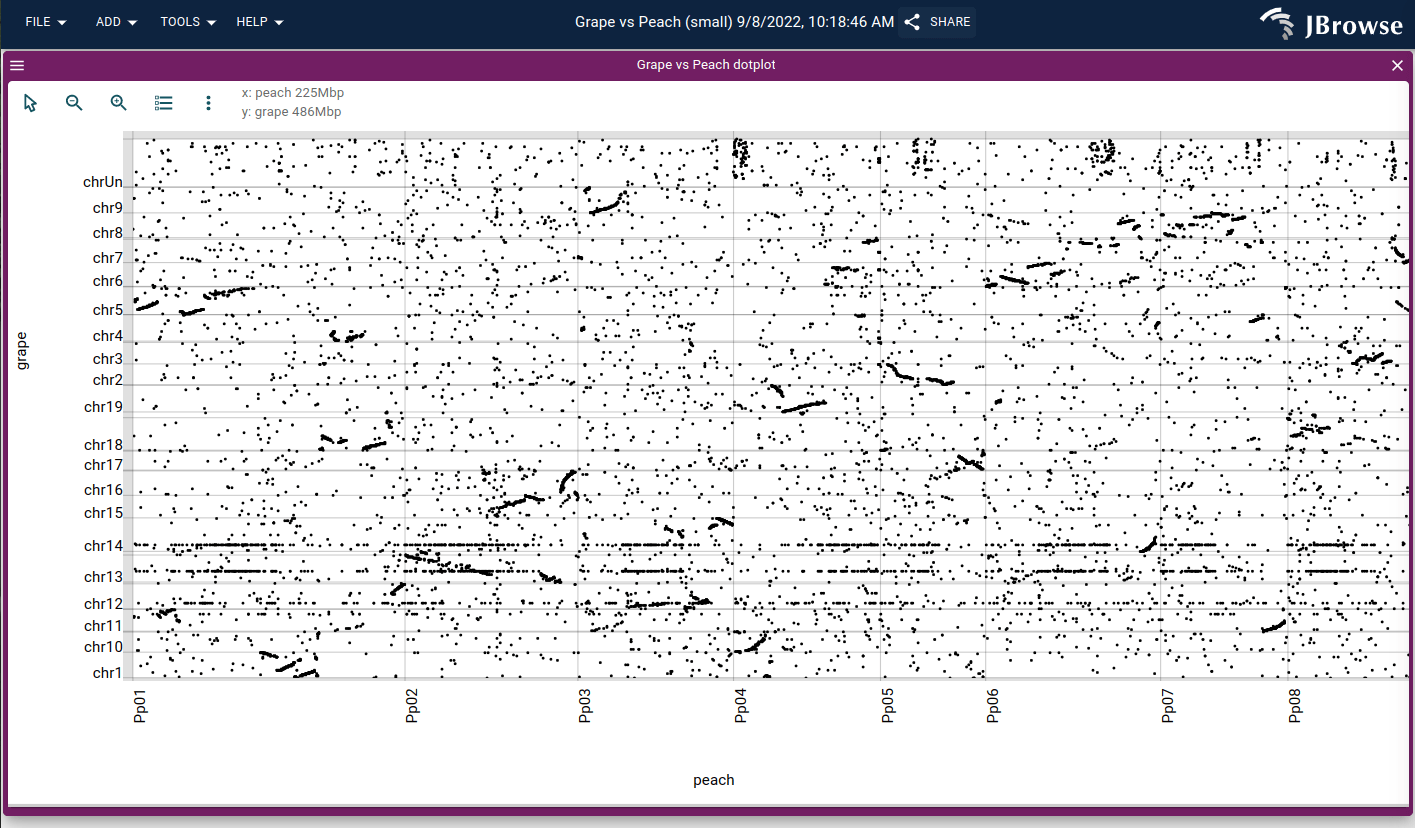
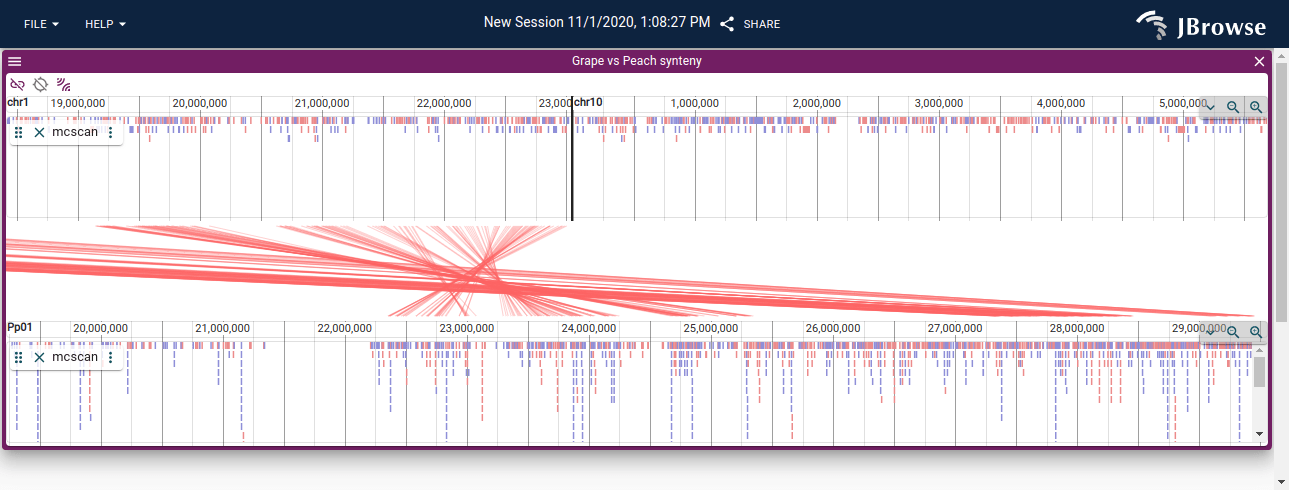
Opening a synteny view from a dotplot view
You can open a synteny view from a dotplot view by selecting a region on the dotplot and clicking "Open linear synteny view", shown below: